An overview of the published plant genomes
(A) The number of plant genomes sequenced up to now (Dec 2023) since the publication of the Arabidopsis thaliana genome in 2000.
(B) Distribution of chromosome-level genome assembleis generated every year.
(C) The top 10 families with the most sequenced genomes in the previous twenty years and the last three years.
(D) Composition of different ploidy for sequenced plant genomes.
(E) The main contributions of different countries to de novo plant genome assembly and the journals for publication in recent six years.

The phylogeny of sequenced genomes and quality metrics for each taxonomic order
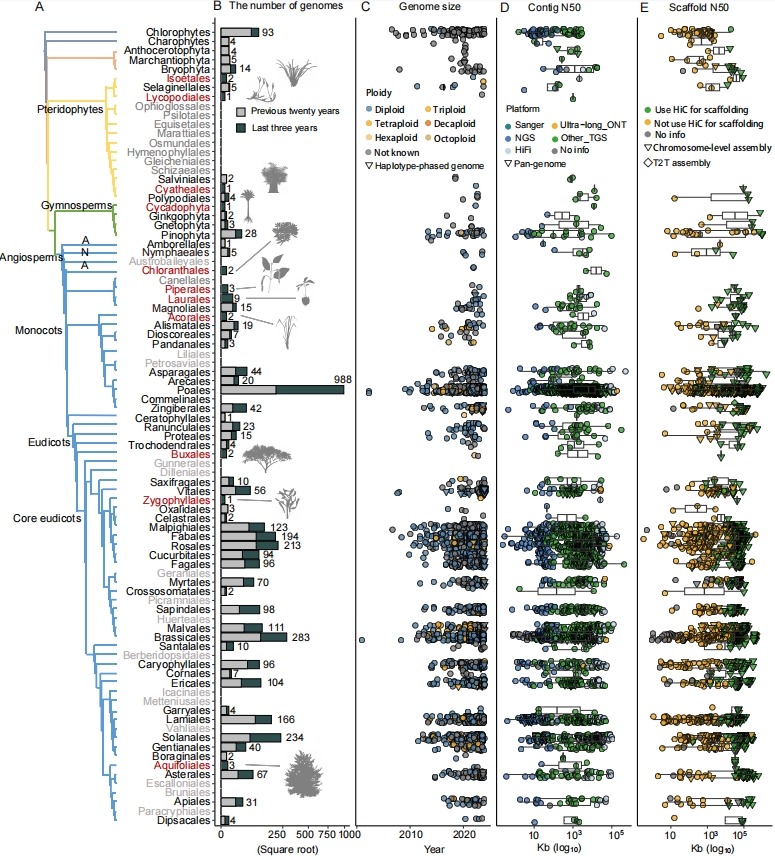
(A) Phylogenetic clades comprised 3,498 sequenced plant genomes across 63 orders. Orders represented in grey font indicated those without any species have been sequenced. Additionally, a comparison between the previous twenty years and the last three years highlights twelve newly included orders in red font.
(B) The number of genomes with publicly available genomes assemblies in the previous twenty years (grey) and the last three years (dark green).
(C) Distribution of genome assembly size for each taxonomic order. Points were coloured by ploidy and shaped as triangles to represent haplotype-phased genomes.
(D) Distribution of contig N50 size for each taxonomic order. Points were coloured by sequencing platform and shaped as triangles to represent pan-genomes.
(E) Distribution of scaffold N50 size for each taxonomic order. Points were coloured by whether HiC data were utilized for scaffolding.

(A) Phylogenetic clades comprised 3,498 sequenced plant genomes across 63 orders. Orders represented in grey font indicated those without any species have been sequenced. Additionally, a comparison between the previous twenty years and the last three years highlights twelve newly included orders in red font.
(B) The number of genomes with publicly available genomes assemblies in the previous twenty years (grey) and the last three years (dark green).
(C) Distribution of genome assembly size for each taxonomic order. Points were coloured by ploidy and shaped as triangles to represent haplotype-phased genomes.
(D) Distribution of contig N50 size for each taxonomic order. Points were coloured by sequencing platform and shaped as triangles to represent pan-genomes.
(E) Distribution of scaffold N50 size for each taxonomic order. Points were coloured by whether HiC data were utilized for scaffolding.

Utilization of sequencing platforms and software employed in assembling process
(A) Contig N50 size across submission date for 2,035 plant genomes is depicted, with points coloured by the types of sequencing platform used. Pan-genome projects are scaled according to the number of genome assemblies for each species. “HiFi” means HiFi reads generated by PacBio sequencing platforms and “ONT” refers to ONT ultra-long reads (reads N50 size ≥100kb); “Other TGS” refers to sequencing technologies that incorporate the third-generation sequencing platforms (HiFi and ONT not included); “NGS” refers to the use of only the second-generation sequencing platforms.
(B) The top 5 software used in three key stages ("contig assembly" in blue, "polishing" in green, and "scaffolding" in yellow) of plant genome assembly, on selected time scales. Software marked with an asterisk is used for assembling chromosomes using Hi-C data.
Reference:Lingjuan Xie#, Xiaojiao Gong#, Kun Yang, Leti Shen, Yujie Huang, Shiyu Zhang, Leti Shen, Yanqing Sun, Dongya Wu, Chuyu Ye, Qian-Hao Zhu, Longjiang Fan*. Technology-enabled great leap in deciphering plant genomes. unpublished.


