Plant single cell lncRNAs
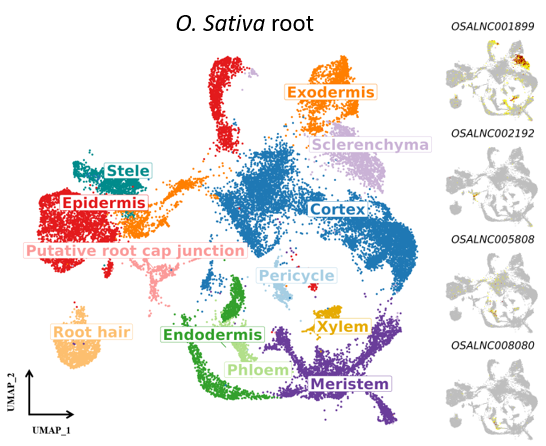
PscLncRNA is a database for plant single-cell lncRNA analysis, which includes four species (Oryza sativa, Arabidopsis thaliana, Zea mays, Solanum lycopersicum). In the latest version, we only focused on intergenic lncRNAs (lincRNAs).
- Example:
- OSALNC001899
- OSALNC002192
- OSALNC005808