Upload
Data upload guidelines for the Orphancereal Omics Database
We warmly invite you to share your data with our database to support orphan cereal crop research! By contributing genome, transcriptome, population, and functional gene data, you help expand the scientific resources available to the research community.Submitting your data is easy! You just need to click on the data type below and follow the prompts to submit the corresponding data
Other Databases
Here we provide commonly used crop genome databases.
Release Note
Release 1.0
2024-9-20
In the first release of the Orphancereal Omics Database, the database is organized into four modules: the Genomes Module stores 217 genome assemblies, which are available for download along with detailed information such as assembly size, contig N50, and related articles. The Transcriptomes Module offers FPKM and TPM data for 6,107 accessions derived from various tissues. The Variants Module contains 45 VCF files with SNPs information from 16,965 samples, 7 of which are sourced from published articles, while the rest are provided by the database. The population-related phenotypic information collected from the literature is also stored in this module. The Functional Genes Module includes detailed information on 147 validated functional genes, covering gene names, associated traits, and relevant literature. The database also features tools like a genome browser (JBrowse) for intuitive exploration of genomic and SNPs information. An online BLAST tool allows users to search for homologous gene sequences across the 28 species’ genomes using any sequence.
(by Fanjing Yang, Xiaojiao Gong, and Chuyu Ye)
Release 1.1
2025-2-28
In the second version of the Orphancereal Omics Database, we have expanded our multi-omics data to include four additional orphan crops: Oryza glaberrima, Triticum spelta, Triticum durum, and Triticum monococcum subsp. monococcum. To enhance data transparency and usability, we have introduced a dedicated methods section on the About page. Additionally, we have incorporated transcriptome sample treatment information and significantly improved the graphical interface's performance in the transcriptome section. Furthermore, we have updated the database's charts to provide more timely and insightful visualizations. In this version, we have also introduced a data upload feature, allowing users to contribute their datasets. This functionality is accessible via the About page, further promoting collaboration and data sharing within the research community.
(by Fanjing Yang, Xiaojiao Gong, and Chuyu Ye)
Data Source
The original data source of the database.
Manual
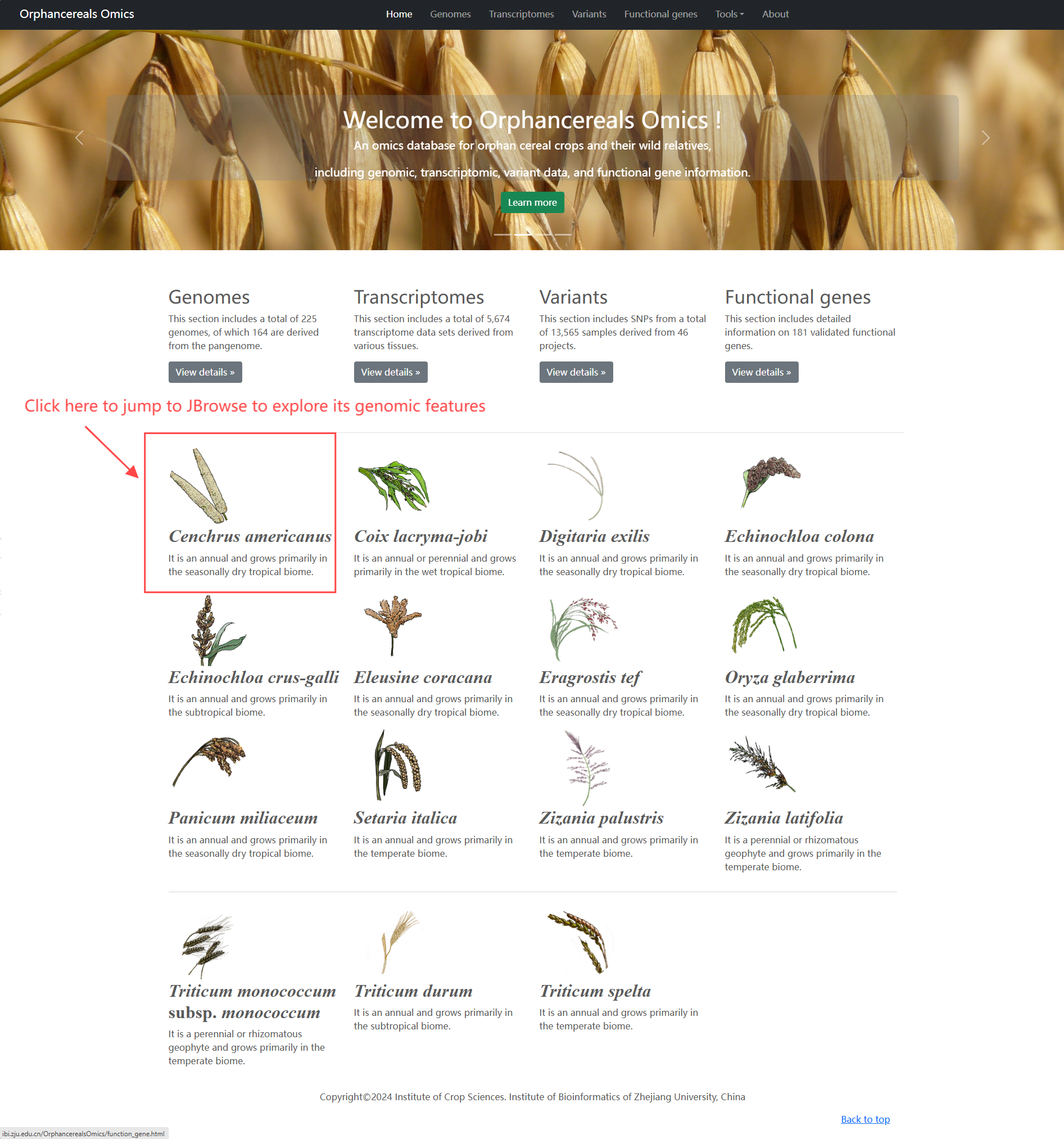
Home page
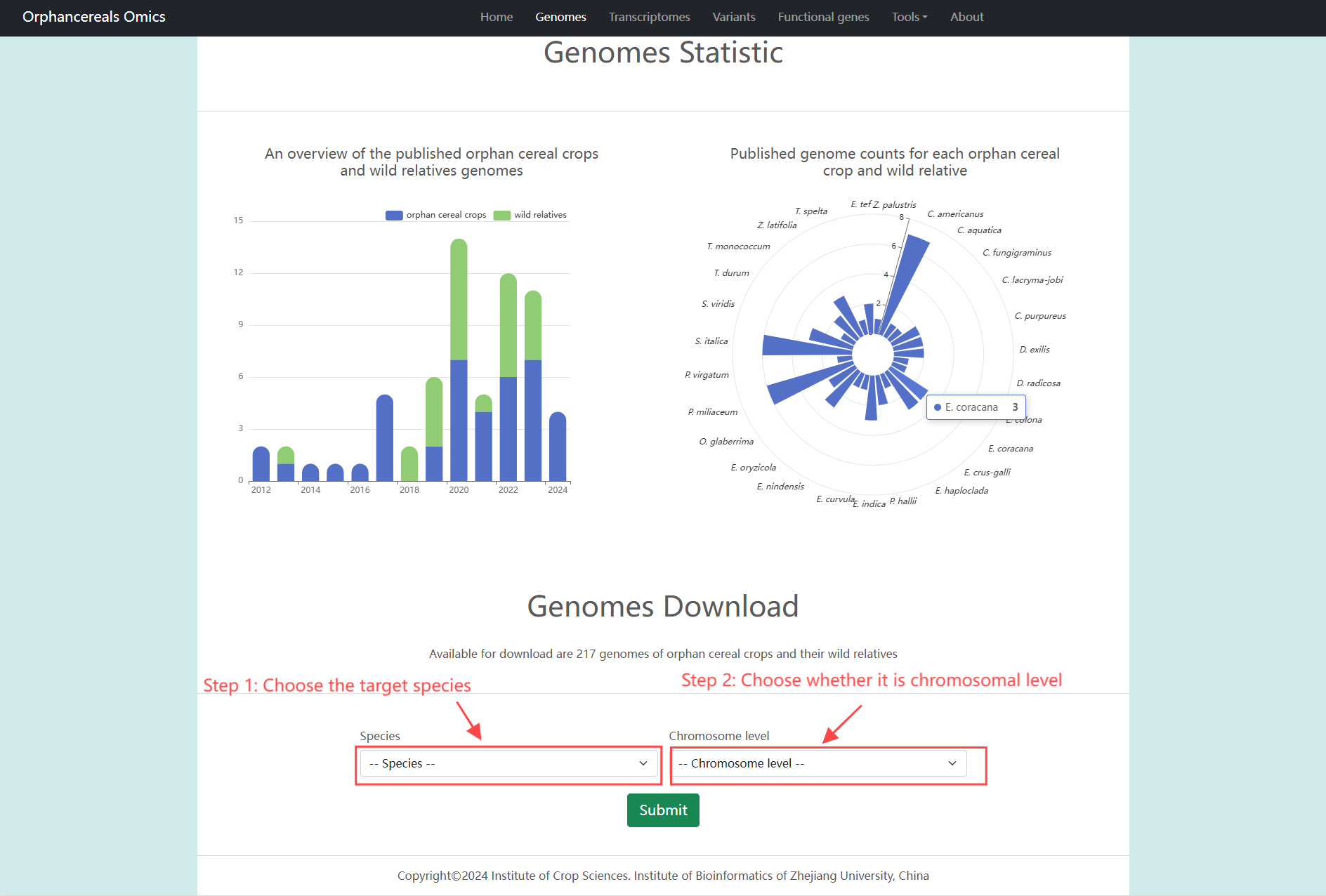
Genomes page
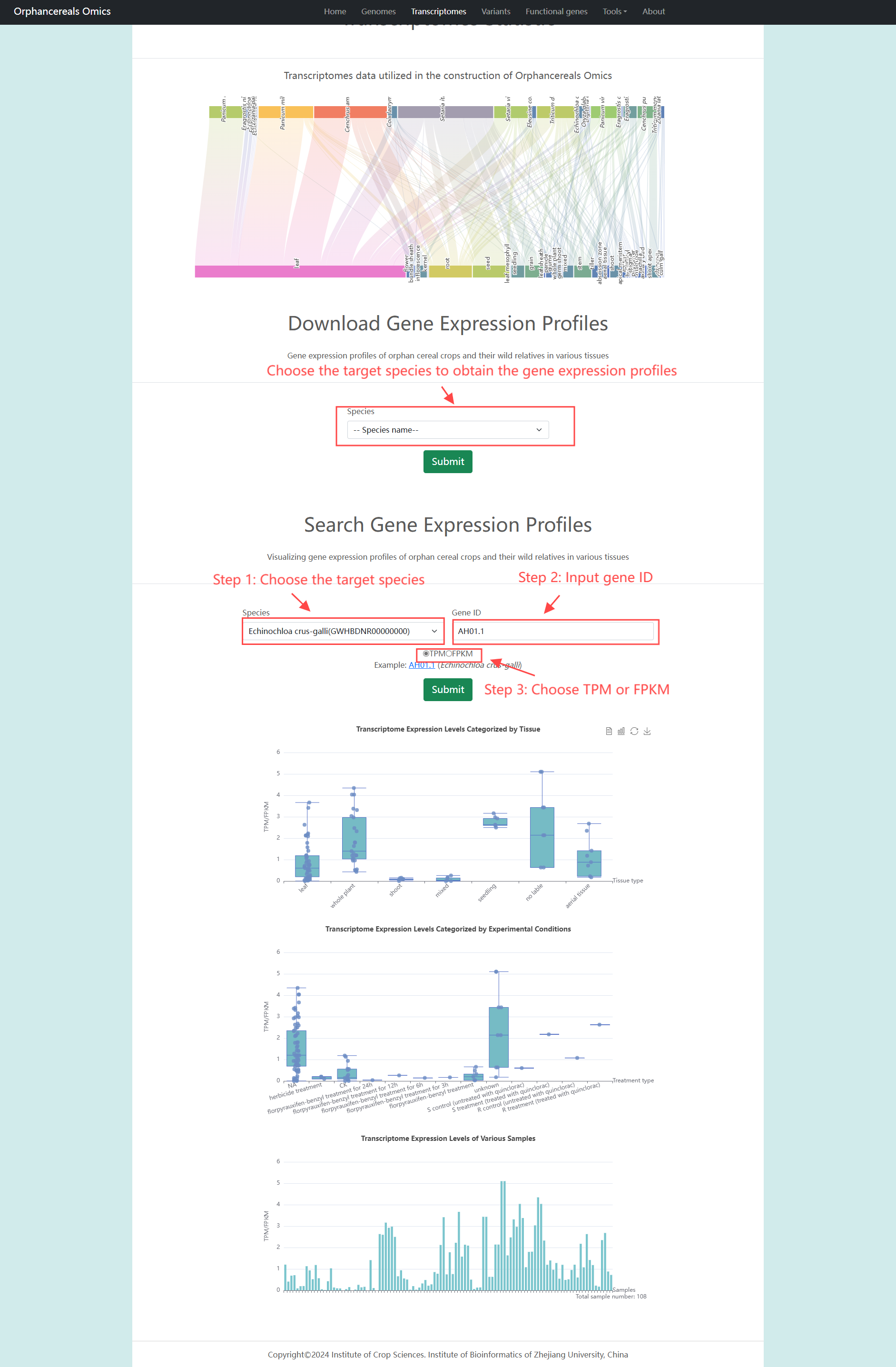
Transcriptomes page
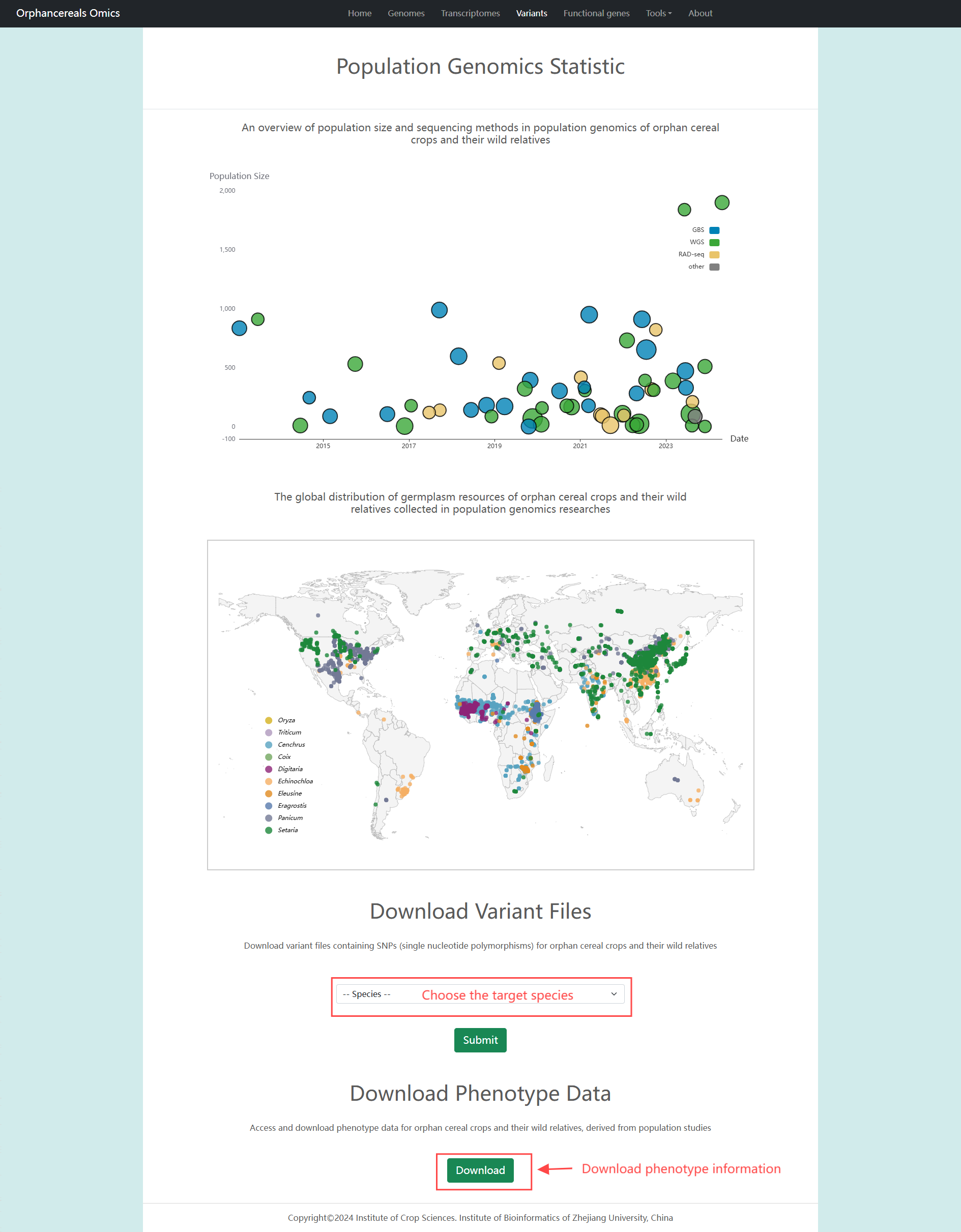
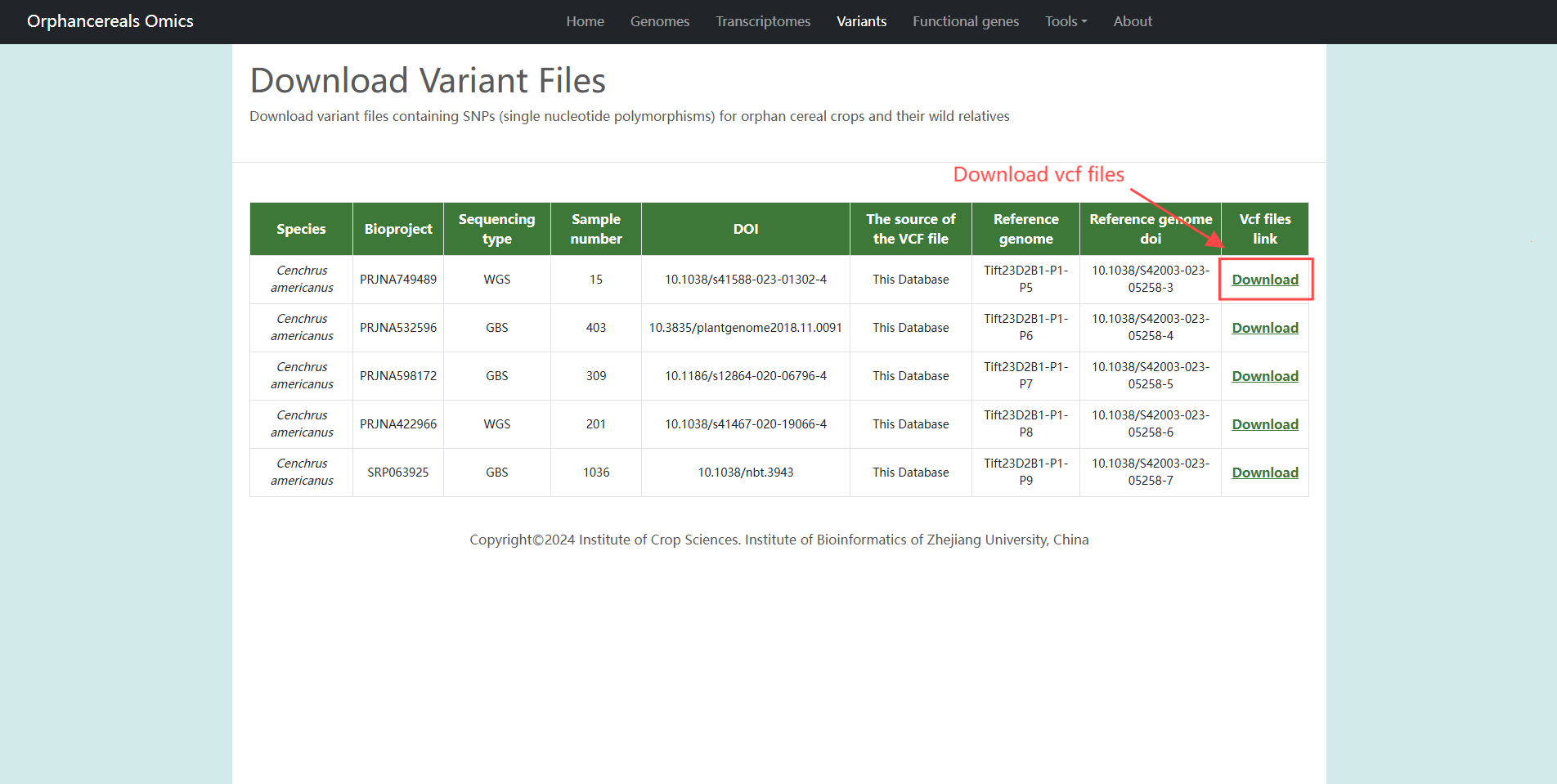
Variants page
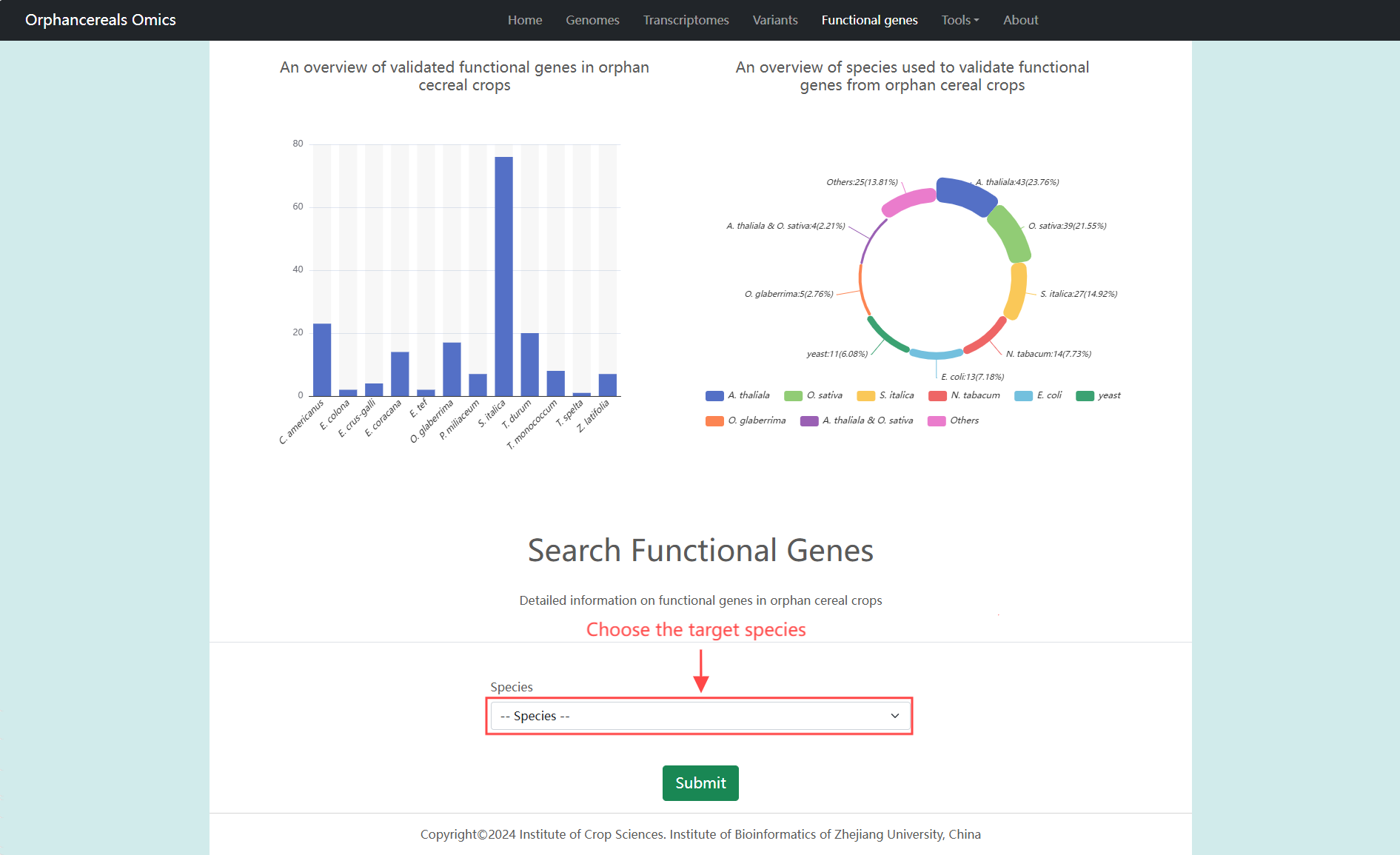
Functional genes page
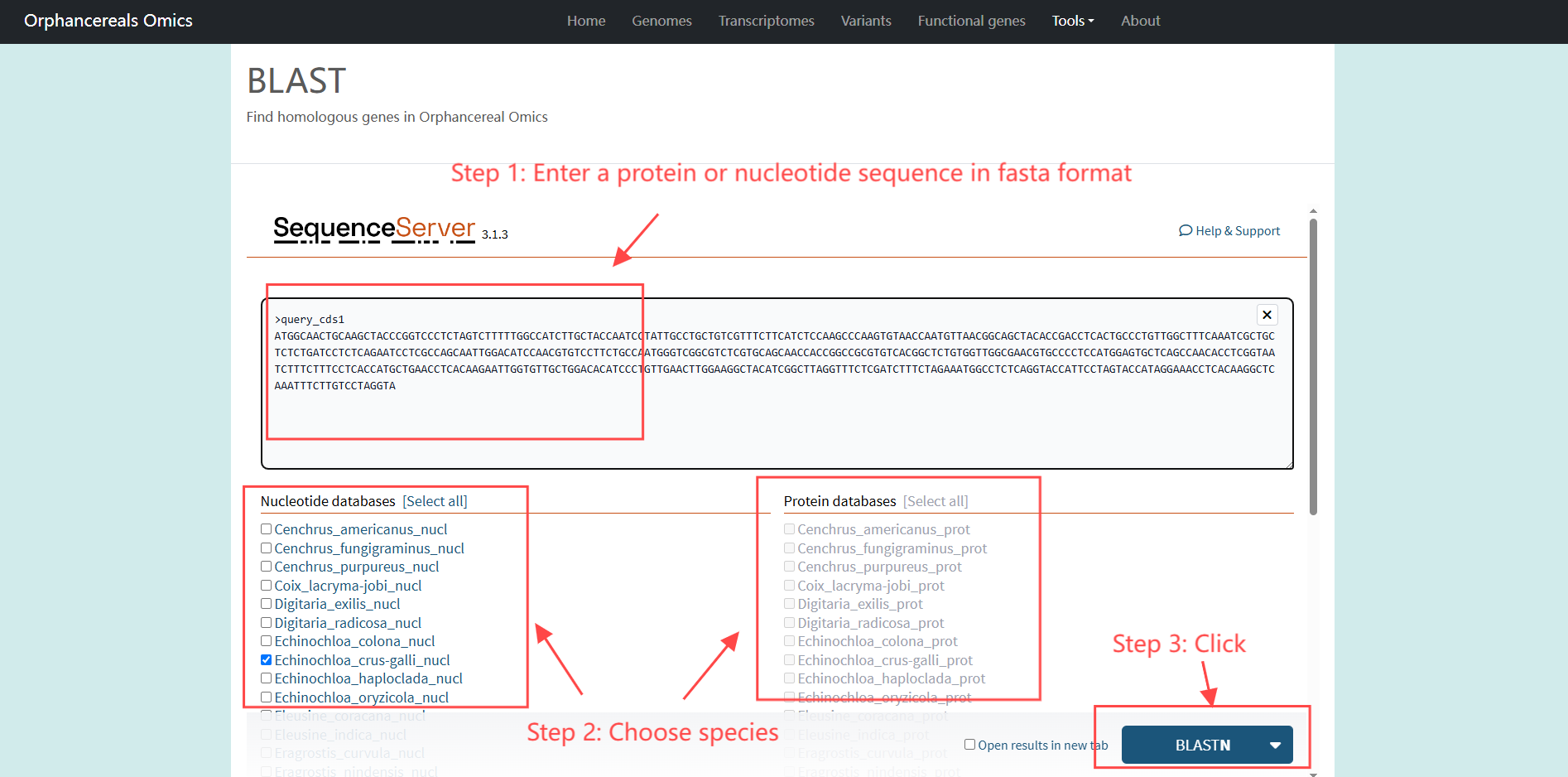
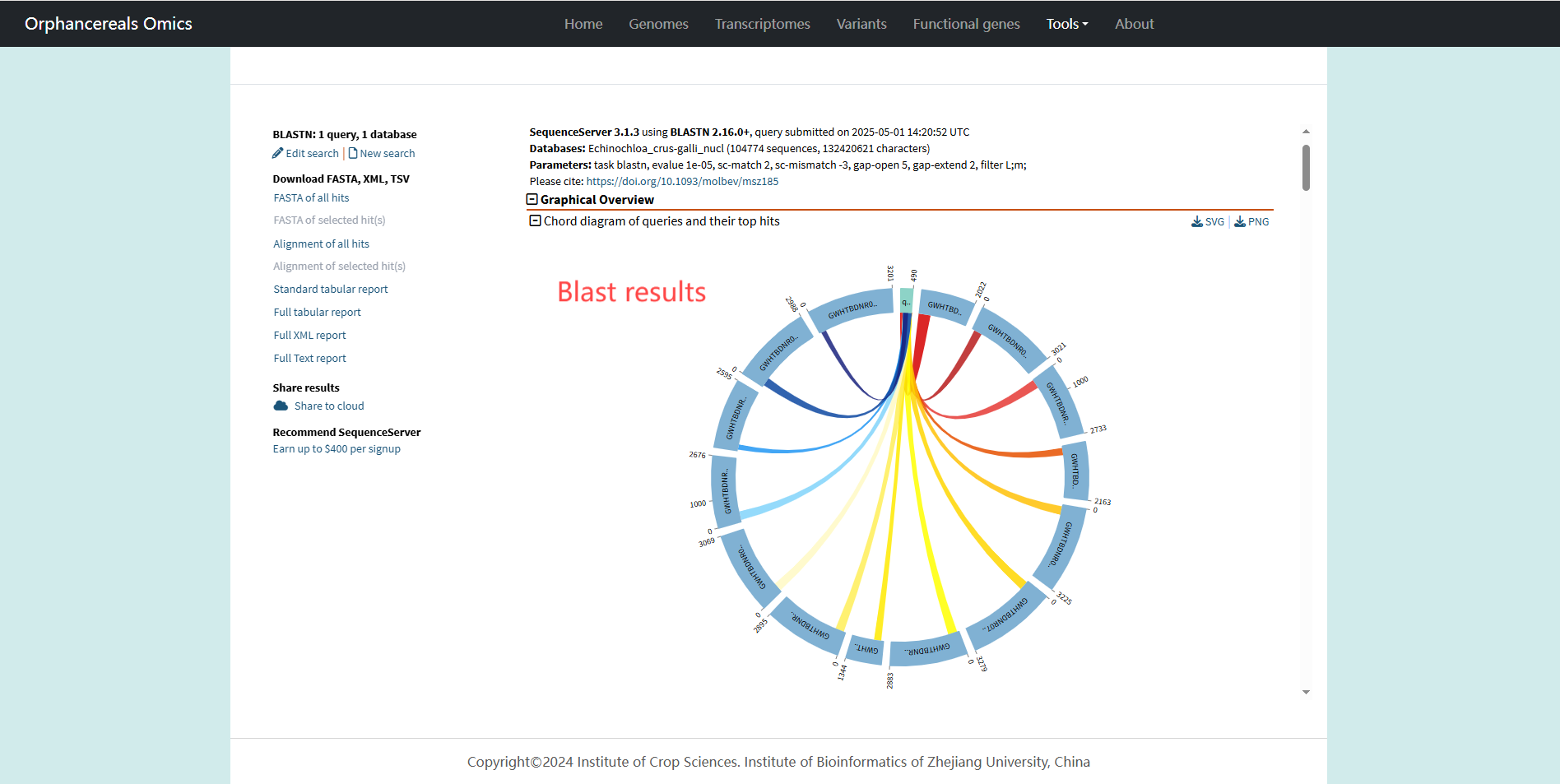
Tools page
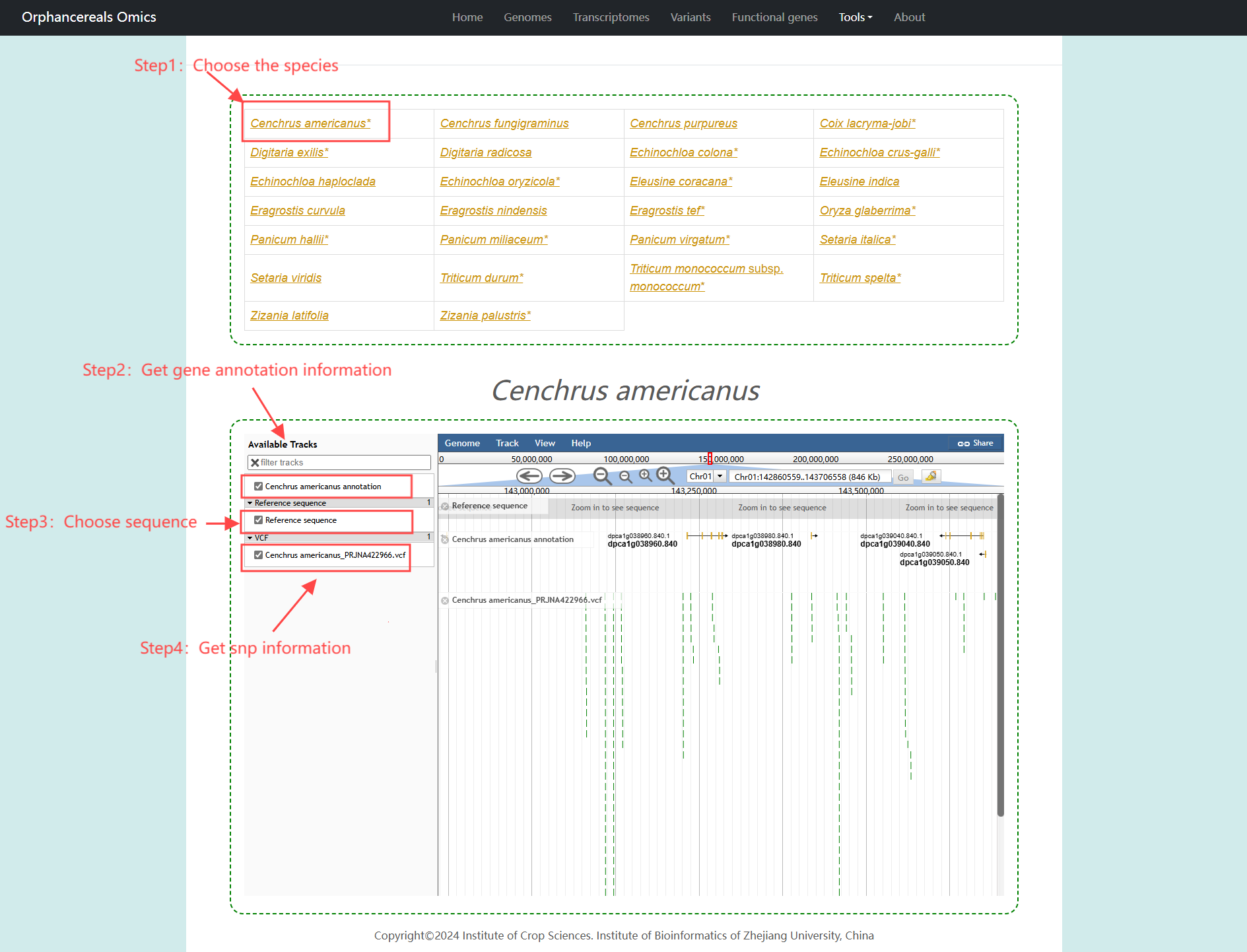
Upload and other databases
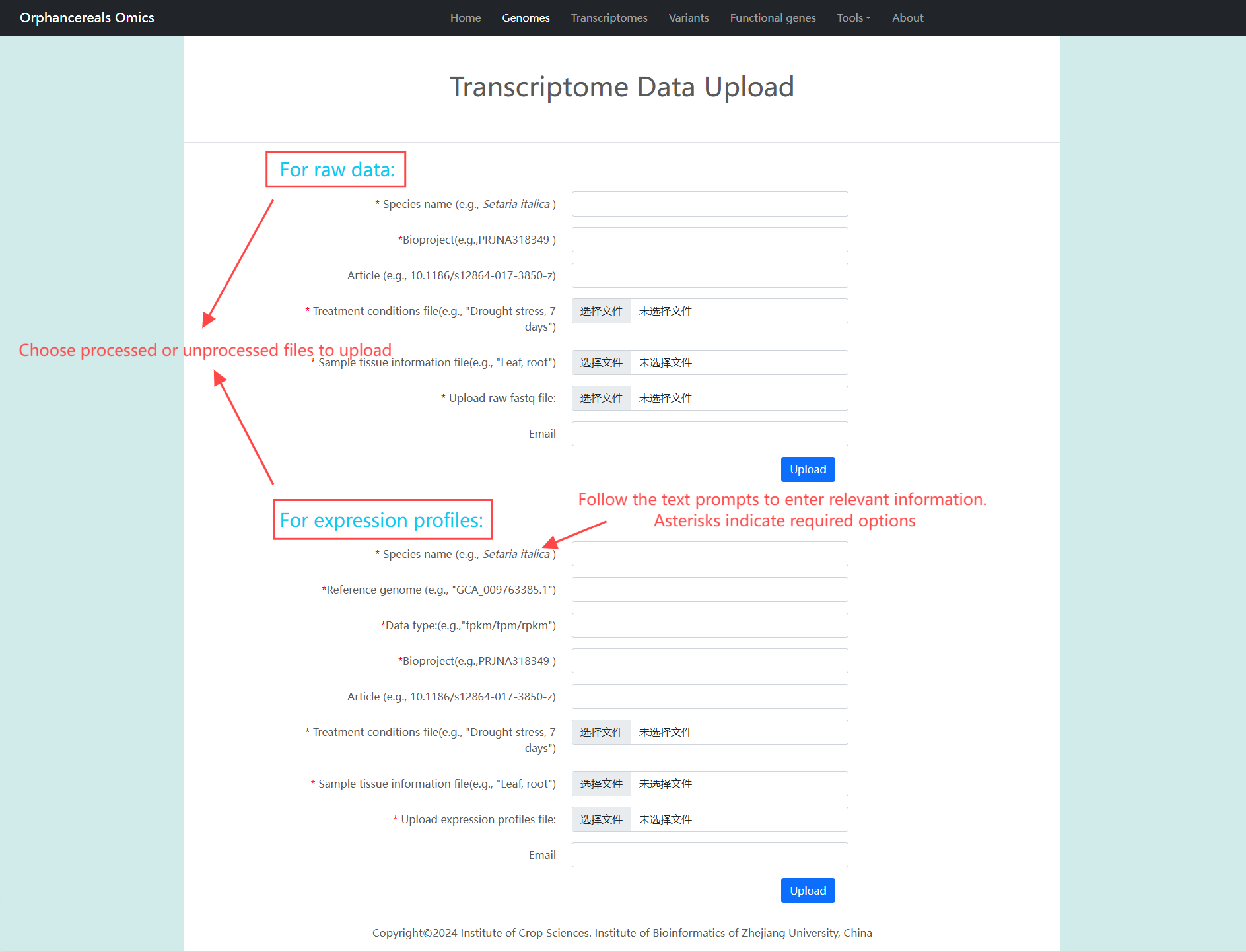
About us
- Institute: Institute of Crop Sciences / Institute of Bioinformatics, Zhejiang University
- Address: Yuhangtang Road, Hangzhou, Zhejiang, China
- E-mail: yecy@zju.edu.cn